Mu-Illumina Gene Identification Service
We invite requests from other groups in the U.S. to use Mu-Illumina sequencing (Williams-Carrier et al, Plant J 2010) to identify Mu-insertions that cosegregate with (putative) Mu-tagged alleles. We can accommodate up to four such requests each year.
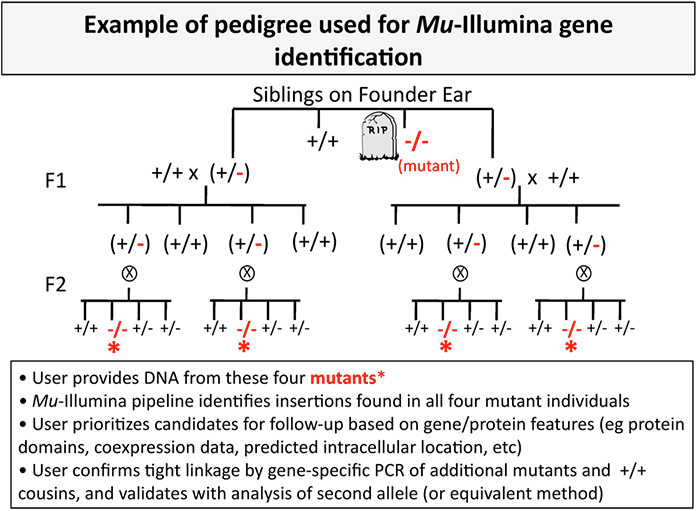
The method requires a pedigree that provides adequate segregation of unlinked insertions from the causal mutation. A two-generation pedigree similar to the one shown below is sufficient.
Our analysis will report a handful of Mu insertions that are tightly linked to the mutation causing the phenotype. Users are responsible for testing candidates by gene-specific PCR, and validation with second alleles, complementation, etc. We recommend use of the POGs2 database as a first stop when evaluating which candidates to prioritize for follow-up.
Our success rate in finding causal mutations with this method is ~85%. Most failures are likely due to alleles that are not caused by Mu and to gaps in the draft maize genome sequence.
Results can be expected within 2-3 months after providing the DNA samples.
Please contact Alice Barkan (abarkan@uoregon.edu) if you are interested in this service.